How to Improve the FAIRness of Biomedical Big Data Research
It’s one of the first things a student is taught in science class: during an experiment, keep track of every step you take. But while the typical school biology experiment is easily documented in a lab book, much of today’s leading research is far more complex. Biomedical experiments now commonly involve massive datasets, often stored in multiple locations, analyzed with a broad array of software tools, while using large, distributed computational resources. Because reproducibility — the ability of an independent researcher to recreate a study and verify its results — is at the heart of the scientific method, it’s critical to find a way to capture even these “big data” workflows.
To address this challenge, members of the scientific community came up with a set of principles and a snappy acronym: Findable, Accessible, Interoperable, and Reusable, or FAIR. By following these guidelines, scientists conducting data and computation-heavy research can share their work in a manner that encourages transparency, interpretability, and reproducibility. In a new study published by PLoS ONE, UChicago CS and Argonne scientists lead a team demonstrating that these goals are both possible and practical, with a collection of low-cost and easy-to-use tools.
“A lot of the great discoveries in genomics are now done using the power of large scale data analysis, and these discoveries have real impact on medicine and on how diseases get treated,” said Ravi Madduri, a UChicago CASE senior scientist and the lead author of the study. “But reproducibility is one of the challenges that science faces in general. When you’re basing your recommendations on a discovery that’s based on a data set that you can’t validate, then it produces a real crisis of confidence.”
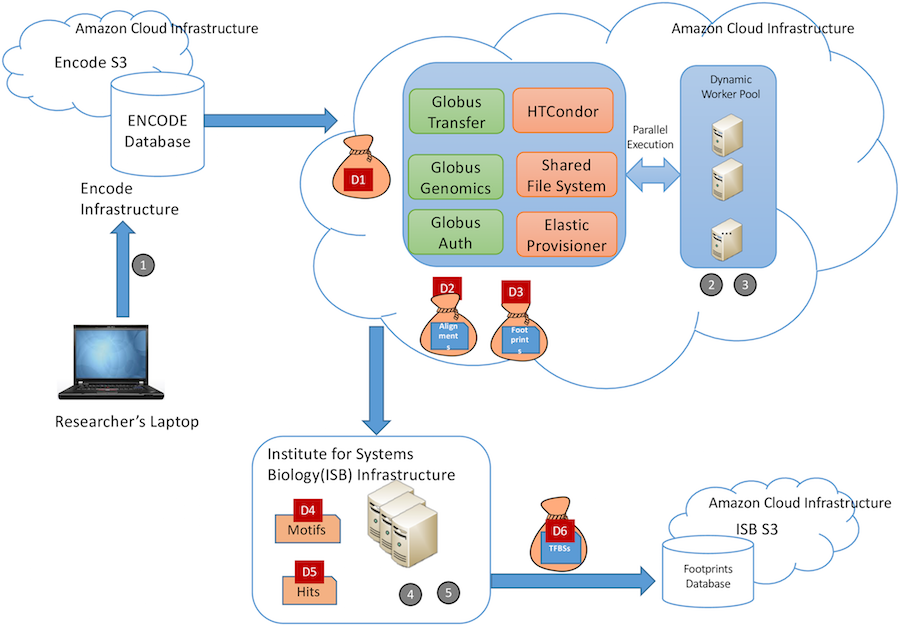
For the study, Madduri and his team (which also included UChicago CS professor Ian Foster, as well as Kyle Chard, Segun Jung, Alexis Rodriguez, and Dinanath Sulakhe of the UChicago/Argonne Globus project), used a case study of the type of big data project that the FAIR principles were meant for. Working with biology researchers from USC, the Institute for Systems Biology in Seattle, and the University of Washington, the team created an “atlas” of transcription factor binding sites using data from the Encyclopedia of DNA Elements (ENCODE), one of the largest and most-used genomic repositories.
“It’s a good example because it is collaborative, team science with researchers from multiple institutions,” Madduri said. “It’s a distributed team working on validating a hypothesis that also has a distributed nature to it.”
The workflow they built involved running nearly 10 terabytes of data through almost 70,000 core hours on cloud computing. Along the way, tools such as BDBag, Minid, Globus, Globus Genomics, and Docker were used to automatically annotate, transfer, analyze, and publish the data in accordance with the FAIR guidelines. Notably, the authors set a high bar for themselves by applying FAIR principles to every step of the research process, from raw source data through intermediate datasets created during analysis steps to the final published product.
“We made a conscious decision to do it in a more methodical fashion to capture all of the raw data that is used, all the analysis that was run, all the results being produced along the way,” Madduri said. “That way, the final results that are generated have a very strong pedigree; we know exactly how this data has been produced, the version of the tools and the parameters used. That boosts a lot of confidence for downstream data reuse.”
To confirm that the process they built was useful and understandable outside of their team, the authors ran a user study to see if other scientists could recreate their work. Happily, 10 out of the 11 participants they selected could run the analysis based on their instructions, while the 11th was foiled by an all-too-familiar problem in data science — difficulties installing the necessary version of R. Because of the user study success, and the team’s commitment to using low-cost and widely-available tools, Madduri hopes that their solution for big data reproducibility will itself be reproducible.
“The approach that we have taken may not translate to everybody, it may only work for a certain type of research, but the tools are generally applicable whatever domain a researcher is working in,” Madduri said. “I hope people take a look at the approach,and think about how those tools can be useful for their own science…I would consider that to be a win.”













