EuroVis Best Paper Awarded to UChicago Diderot Team
New technologies can produce richer scientific images than ever before, providing high-resolution, three-dimensional views of cellular structures, intricate brain networks, and distant stars. But these methods also create new data challenges for scientists who need to extract specific information from complex structures and terabyte-sized files.
In 2011, University of Chicago computer scientists Gordon Kindlmann and John Reppy began building Diderot, a new programming language specifically designed for the visualization and analysis of this class of image data. Earlier this month, a new approach using Diderot to render and discover features of interest from 3D structures was presented at the prestigious EuroVis conference, receiving the award for Best Paper.
“This paper presents a novel, concise, and systematic mathematical basis for unifying approaches to describe features in 3D volumes,” the conference wrote on its website. “The paper is well-written with an excellent and complete view on previous work and offers a demonstrable improvement in the feature detection process which should have a lasting impact on the discipline.
The new functionality announced at EuroVis reflects the original principles of Diderot, allowing scientists to copy mathematical notation underlying their target features into legible, transparent, and concise code.
“The premise for Diderot is that the world is changing, the kinds of data that we gather are changing, and the kinds of questions that we ask about data are changing,” Kindlmann said. “The human cost of implementing new methods becomes a bigger bottleneck than the computational expense of executing existing implementations. So the goal of Diderot is to simplify the path from idea to working code.”
Typically, when a scientist captures new images, they next look for specific areas of interest, such as a particular cell type or landmark. To automate this search, researchers select a geometric signature — for example, an area of relative brightness or a characteristic shape — that can be expressed as a mathematical equation and applied to the data to find all of its instances.
At the core of the new paper are two new programs in Diderot that allow users to conduct two critical pieces of this process, direct volume rendering and particle-based feature sampling, by adding as little as one line of code. To find a particular feature, users only need to derive an expression for the “Newton step” for the feature, which is then written in Diderot to complete the programs.
“With a little bit of work at the whiteboard, I can figure out the Newton step, plug that into the code, and now I have a way of looking at the feature and a way of obtaining its geometry for further analysis,” Kindlmann said.
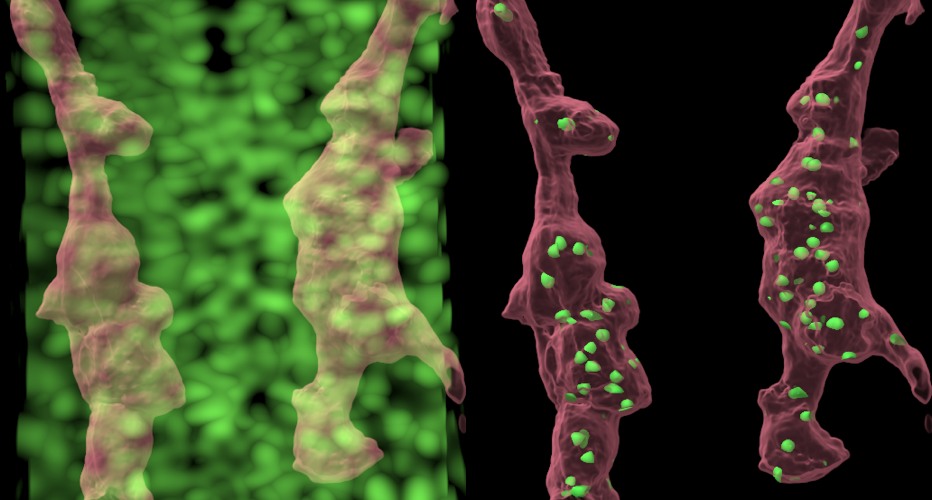
The applications of this approach are manyfold. In the paper, the authors present a collaboration with Prof. Victoria Prince and Ana Beiriger from the UChicago Department of Organismal Biology and Anatomy, using fluorescent microscopy images of developing zebrafish neurons. With the equation for finding local maxima — areas of elevated brightness — Diderot produces much clearer visualizations, allowing the scientists to closely track the movement of fluorescent-tagged nuclei [see before and after images on right].
Another case study examines the use of several different features to reveal the anatomical structure of human brains from diffusion MRI images. Scientists can use these features to isolate major axonal pathways of the brain or identify specific landmarks that could improve group studies over many individuals.
“There's a dozen ways of trying to pull structure out of a diffusion tensor field that may or may not have reliable anatomical meaning. And now it's a whole lot easier to explore those approaches,” Kindlmann said. “It changes the implementation cost of exploring these features from weeks of work to minutes of work.”
In addition to Kindlmann and Reppy, study co-authors include Charisee Chiw and Tri Huynh of UChicago and Attila Gyulassy and Peer-Timo Bremer of the University of Utah.